Sage search engine#
Sage [LAZ2023] is an open source, high-performance, and freely available proteomics search engines. Scalable and cloud-ready, Sage matches the performance of state-of-the-art software tools while running an order of magnitude faster.
Note
In quantms, Sage is in the middle of the pack in terms of number of identified PSMs, CPU usage and speed compared to MSGF+ and Comet.
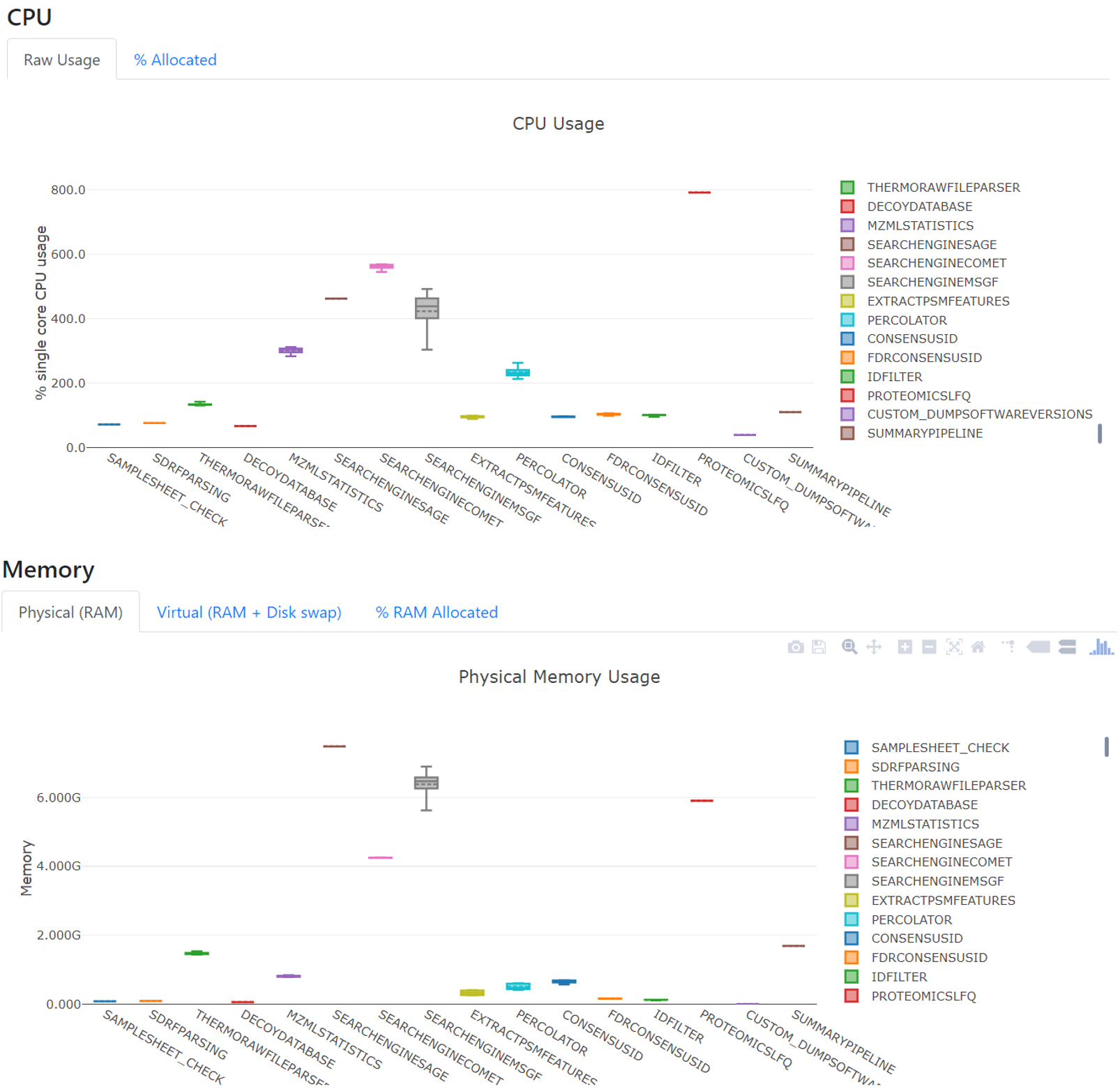
In quantms, PSMs are exported from the search engine into .idXML (read more Formats in quantms) without filtering for the re-scoring (see Peptide identification from fragment spectra) step with percolator. The pipeline stores these original file results in the result folder under searchenginesage.
References#
Lazear MR. Sage: An Open-Source Tool for Fast Proteomics Searching and Quantification at Scale. J Proteome Res. 2023 Nov 3;22(11):3652-3659. doi: 10.1021/acs.jproteome.3c00486. Epub 2023 Oct 11. PMID: 37819886.