MS-GF+ peptide search engine#
MS-GF+: MS-GF+ (aka MSGF+ or MSGFPlus) [KIM2014] performs peptide identification by scoring MS/MS spectra against peptides derived from a protein sequence database. MSG-GF+ is released under specific License from California University.
Note
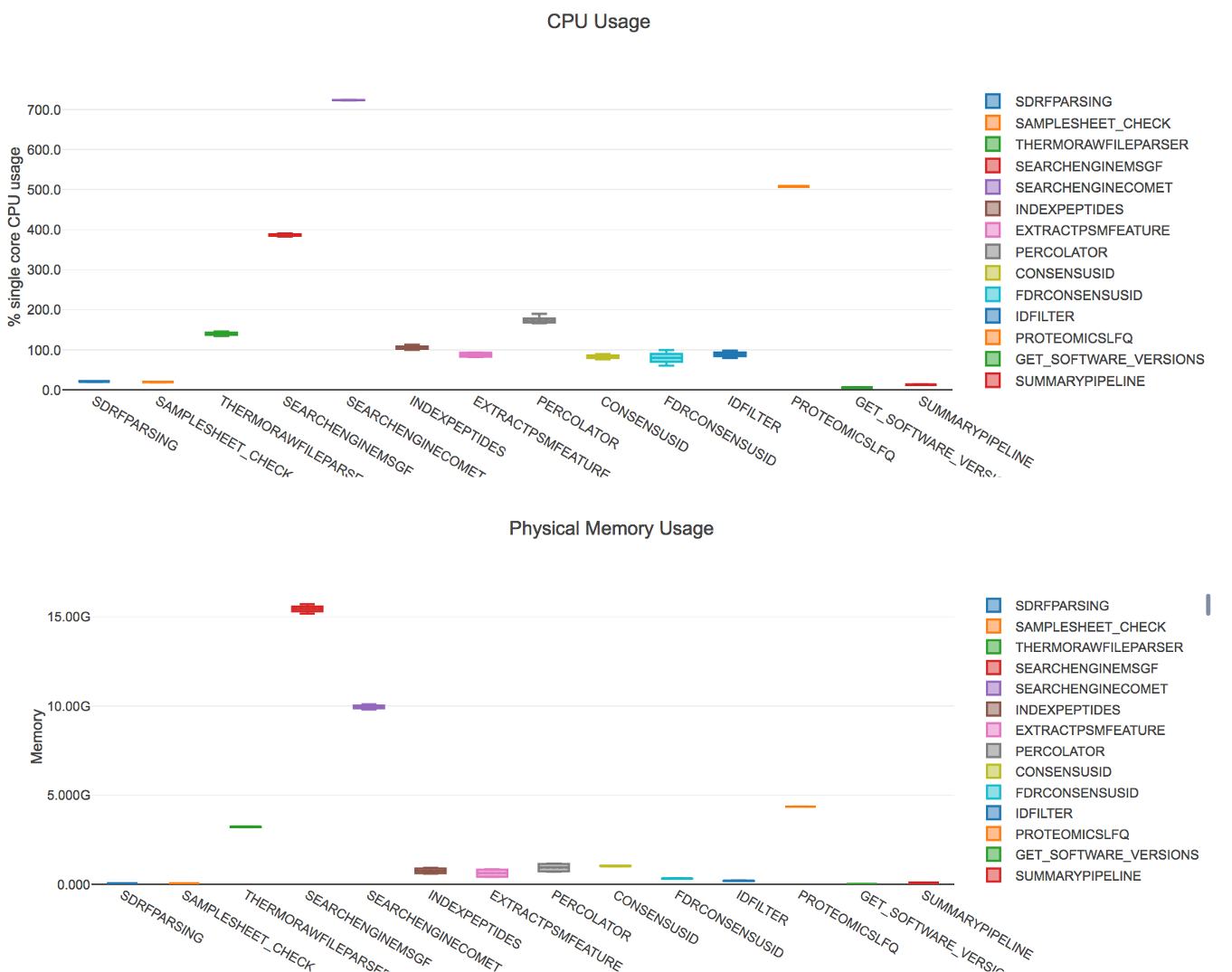
In quantms, msgf+ has proved to identified more than 15% PSMs than Comet search engine. However, users of cloud infrastructure needs to consider that msgf+ may slow the data processing and consume more memory than Comet search engine.
In quantms, PSMs are exported from the search engine into .idXML (read more Formats in quantms) without filtering for the re-scoring (see Peptide identification from fragment spectra) step with percolator. The pipeline stores these original file results in the result folder under searchenginemsgf.
References#
Sangtae Kim and Pavel A Pevzner. Nat Commun. MS-GF+ makes progress towards a universal database search tool for proteomics. 2014 Oct 31; 5:5277. doi: 10.1038/ncomms6277. PubMed ID 25358478